This is a dataset for evaluating entailment detection between dependency graph fragments. For more information, please refer to the following paper:
Unsupervised Entailment Detection between Dependency Graph Fragments In Proceedings of the 2011 Workshop on Biomedical Natural Language Processing (BioNLP-11). Portland, United States, 2011
Download
Dataset of Entailment Between Dependency Graph Fragments
General definitions
We define a fragment in the following way:
Definition 1. A fragment is any connected subgraph of a directed dependency graph containing one or more words and the grammatical relations between them.
This definition is intended to allow extraction of a wide variety of fragments from a dependency tree or graph representation of sentences found using any appropriate parser capable of returning such output (e.g. Kübler et al., 2009). The definition covers single- or multi-word constituents functioning as dependents (e.g. sites, putative binding sites), text adjuncts (in the cell wall), single- or multi-word predicates (* binds to receptors in the airways) and relations (* binds and activates *) including ones with ‘internal’ dependent slots (* inhibits * at *), some of which may be fixed in the fragment (* induces autophosphorylation of * in * cells), and also full sentences. An example dependency graph and some selected fragments can be seen in Figure 1.
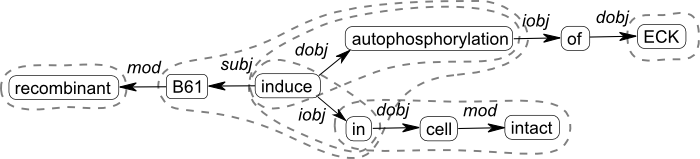 Figure 1. Dependency graph for the sentence: Recombinant B61 induces autophosphorylation of ECK in intact cells. Some interesting fragments are marked by dotted lines.
Figure 1. Dependency graph for the sentence: Recombinant B61 induces autophosphorylation of ECK in intact cells. Some interesting fragments are marked by dotted lines.
Our aim is to detect semantically similar fragments which can be substituted for each other in text, resulting in more general or more specific versions of the same proposition. This kind of similarity can be thought of as an entailment relation and we define entailment between two fragments as follows:
Definition 2. Fragment A entails fragment B (A → B) if A can be replaced by B in sentence S and the resulting sentence S’ can be entailed from the original one (S → S’).
This also requires estimating entailment relations between sentences, for which we use the definition established by Bar-Haim et al. (2006):
Definition 3. Text T entails hypothesis H (T → H) if, typically, a human reading T would infer that H is most likely true.
Dataset construction
A ‘pilot’ dataset was created to evaluate different entailment detection methods between fragments. In order to look for valid entailment examples, 1000 biomedical papers from the BioMed Central full-text corpus were randomly chosen and analysed. We hypothesised that two very similar sentences originating from the same paper are likely to be more and less general versions of the same proposition. First, the similarities between all sentences in a single paper were calculated using a bag-of-words approach. Then, ten of the most similar but nonidentical sentence pairs from each paper were presented for manual review and 150 fragment pairs were created based on these sentences, 100 of which were selected for the final set.
Two annotators assigned a relation type to candidate pairs based on how well one fragment can be substituted for the other in text while preserving meaning (A = B, A → B, A ← B or A ≠ B). Cohen’s Kappa between the annotators was 0.88, indicating very high agreement. Instances with disagreement were then reviewed and replaced for the final dataset. Each fragment pair has two binary entailment decisions (one in either direction) and the set is evenly balanced, containing 100 entailment and 100 nonentailment relations. An example sentence with the first fragment is also included. Fragment sizes range from 1 to 20 words, with the average of 2.86 words.
Dataset format
Each entailment pair consists of 7 lines:
- Example sentence using the first of the two fragments
- The first fragment
- Relation between fragments
- The second fragment
The relation between the fragments is one of the following:
- -> : The first fragment entails the second fragment
- <- : The second fragment entails the first fragment
- == : Both fragments entail each other
- != : No entailment relation between the fragments